Quick Guide¶
Introduction¶
epistasis is a Python library that includes models to estimate statistical, high-order epistasis in genotype-phenotype maps. Using this library, you can
Decompose genotype-phenotype maps into high-order epistatic interactions
Find nonlinear scales in the genotype-phenotype map
Calculate the contributions of different epistatic orders and
Estimate the uncertainty in the epistatic coefficients and
For more information about the epistasis models in this library, see our Genetics paper:
Simple Example¶
Follow these five steps for all epistasis models in this library:
Import a model. There many models available in the
epistasis.modelsmodule. See the full list in the next section.
from epistasis.models import EpistasisLinearRegression
Initialize a model. Set the order, choose the type of model (see Anatomy of an epistasis model for more info), and set any other parameters in the model.
model = EpistasisLinearRegression(order=3, model_type='global')
Add some data. There are three basic ways to do this. 1. Pass data directly to the epistasis model using the
add_datamethod. 2. Read data from a separate file using one of theread_methods. 3. (The best option.) load data into a GenotypePhenotypeMap object from the GPMap library and add it to the epistasis model.
from gpmap import GenotypePhenotypeMap
datafile = 'data.csv'
gpm = GenotypePhenotypeMap.read_csv(datafile)
# Add the data.
model.add_gpm(gpm)
# model now has a `gpm` attribute.
Fit the model. Each model has a simple fit method. Call this to estimate epistatic coefficients. The results are stored the
epistasisattribute.
# Call fit method
model.fit()
# model now has an ``epistasis`` attribute
Plot the results. The epistasis library has a
pyplotmodule (powered by matplotlib) with a few builtin plotting functions.
from epistasis.pyplot import plot_coefs
fig, ax = plot_coefs(model.epistasis.sites, model.epistasis.values)
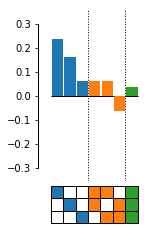
Install and dependencies¶
For users¶
This library is now available on PyPi, so it can be installed using pip.
pip install epistasis
For developers¶
For the latest version of the package, you can also clone from Github and install a development version using pip.
git clone https://github.com/harmslab/epistasis
cd epistasis
pip install -e .
Dependencies¶
The following dependencies are required for the epistasis package.
gpmap: Module for constructing powerful genotype-phenotype map python data-structures.
Scikit-learn: Simple to use machine-learning API.
Numpy: Python’s array manipulation package.
Scipy: Efficient scientific array manipulations and fitting.
Pandas: High-performance, easy-to-use data structures and data analysis tools.
There are also some additional dependencies for extra features included in the package.
matplotlib: Python plotting API.
ipython: interactive python kernel.
jupyter notebook: interactive notebook application for running python kernels interactively.
ipywidgets: interactive widgets in python.
Running tests¶
The epistasis package comes with a suite of tests. Running the tests require pytest, so make sure it is installed.
pip install -U pytest
Once pytest is installed, run the tests from the base directory of the epistasis package using the following command.
pytest